

- #COUNT NUCLEI IMAGEJ HOW TO#
- #COUNT NUCLEI IMAGEJ SOFTWARE#
Select Plugins>Itcn>Itcn, a dialog will appear.Select Edit>Invert so that you have dark signals on a white background.Convert the image to 8-bit using Image>Type>8-bit.Open an image before you start the plugin.
#COUNT NUCLEI IMAGEJ HOW TO#
You can also refer to this video on Youtube for how to use the original version of ITCN. Go to that folder and there will be a plugins subfolder.
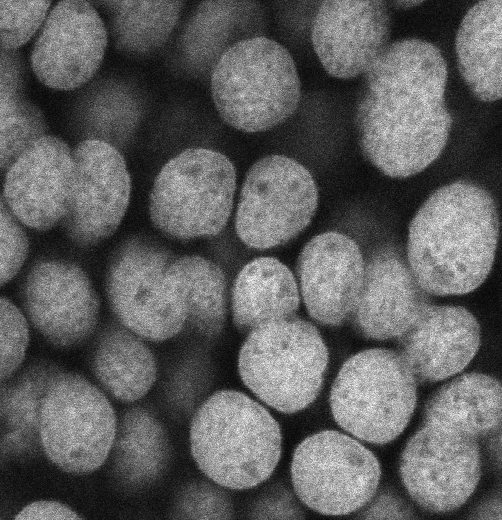
Open the software, CLOSE ALL IMAGES and select Image>Show info.

#COUNT NUCLEI IMAGEJ SOFTWARE#
Open the software and select File>Show Folder>Plugins. There will be a new Plugins>Itcn menu path. Installationĭownload Itcn.zip from this Github directory, unzip, and place it under the ImageJ or Fiji plugins folder, restart ImageJ or Fiji. Here I improved the original plugin a little bit so that it could be recorded by and used in ImageJ macro scripts. However, because the plugin is developed many years ago, it is no longer maintained and no detailed instructions could be found on the internet.

It could also be downloaded from the developer's web page (This version actually works better). The original plugin is developed by the Center for Bio-image Informatics at UC Santa Barbara, and released on the ImageJ website. This is a modified version of the ImageJ plugin ITCN.
Circularity fields: Does the same as above but for circularity.ImageJ Nuclei Counter for Fluorescent Images The modified ImageJ plugin ITCN. Pressing the keyboard key "Enter" in any of these input fields will delete the previously saved *-RoiSet.zip and recount the image with the new settings, saving a new *-RoiSet.zip when the image is closed or replaced by "Open next". Size fields: Allows setting the minimum and maximum limits for the size of colonies that the user wants to detect and count. About: Shows the "About Colony Counter" message window. Delete colony: Activates/Deactivates the "Show all" function of the ROI Manager, allowing selection of a colony by clicking on it's number on the image, and then deleting it by pressing the keyboard letter "d". The last value in the "Results" table and the *-RoiSet.zip file are updated. Add colony: Adds the currently selected area as a colony allowing correcting the automatic count. Count folder: Counts and/or loads the *-RoiSet.zip for all images in the current folder, and replaces the current "Results" table with the new results. Open next: Replaces the current image with the next one in the same folder and do the same as "Open image". The number of colonies is added in a new row to the "Results" table. Open image: Opens an image and automatically counts it's colonies, or simply loads and display the saved *-RoiSet.zip from a previous count. The plugin's interface is simple, on the left are the functions buttons, on the right the input fields for the settings.Įxcept for the "Delete mode" and the "About" buttons, all the others have a keyboard shortcut (visible inside square brackets on the button's name) that can be used when the mouse cursor is placed on the image. When the image is closed, a file named *-RoiSet.zip is automatically saved in the current folder, so that the next time the same image is opened, the plugin won't have to recount that image and can simply load the results from the *-RoiSet.zip. The obtained values are displayed in a "Results" table and can be saved in *.xls file format. This plugin was created to count bacteria colonies from agar plates images. Or subfolder, restart ImageJ, and there will be a new "Colony Counter" command in the Plugins menu or submenu. Included in Colony_Counter.jar, and is released under the GNU General Public License.ĭownload Colony_Counter.jar to the plugins folder, University of Lisbon, Faculty of Sciences







 0 kommentar(er)
0 kommentar(er)
